Emukit and Experimental Design
Simulation System
Monolithic System
Service Oriented Architecture
Experiment, Analyze, Design
A Vision
We don’t know what science we’ll want to do in five years’ time, but we won’t want slower experiments, we won’t want more expensive experiments and we won’t want a narrower selection of experiments.
What do we want?
- Faster, cheaper and more diverse experiments.
- Better ecosystems for experimentation.
- Data oriented architectures.
- Data maturity assessments.
- Data readiness levels.
Packing Problems

Packing Problems
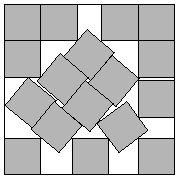
Packing Problems
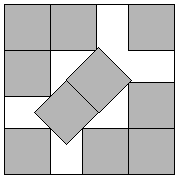
Modelling with a Function
- What if question of interest is simple?
- For example in packing problem: what is minimum side length?
Erich Friedman Packing Data
Erich Friedman Packing Data: Squares in Squares
Gaussian Process Fit
Erich Friedman Packing Data GP
Statistical Emulation
Emulation
Emulation
Emulation
Emulation
Emulation
GPy: A Gaussian Process Framework in Python
GPy: A Gaussian Process Framework in Python
- BSD Licensed software base.
- Wide availability of libraries, ‘modern’ scripting language.
- Allows us to set projects to undergraduates in Comp Sci that use GPs.
- Available through GitHub https://github.com/SheffieldML/GPy
- Reproducible Research with Jupyter Notebook.
Features
- Probabilistic-style programming (specify the model, not the algorithm).
- Non-Gaussian likelihoods.
- Multivariate outputs.
- Dimensionality reduction.
- Approximations for large data sets.
GPy Tutorial
Covariance Functions
\[ k(\mathbf{ x}, \mathbf{ x}^\prime) = \alpha \exp\left(-\frac{\left\Vert \mathbf{ x}- \mathbf{ x}^\prime \right\Vert_2^2}{2\ell^2}\right), \]
Kernel Output
Covariance Functions in GPy
- Includes a range of covariance functions
- E.g. Matern family, Brownian motion, periodic, linear etc.
- Can define new covariances
Combining Covariance Functions in GPy
Multiplication
A Gaussian Process Regression Model
\[ f( x) = − \cos(\pi x) + \sin(4\pi x) \]
\[ y(x) = f(x) + \epsilon, \]
Noisy Sine
GP Fit to Noisy Sine
Covariance Function Parameter Estimation
GPy and Emulation
\[ f(\mathbf{ x}) = a(x_2 - bx_1^2 + cx_1 - r)^2 + s(1-t \cos(x_1)) + s \]
Computation of \(\left\langle f(U)\right\rangle\)
- Compute the expectation of the Branin function.
- Sample at 25 points on a grid.
- Compute the mean (Reiemann sum approximation).
Emulate and do MC Samples
Alternatively, we can fit a GP model and compute the integral of the best predictor by Monte Carlo sampling.
Branin Function Fit
Uncertainty Quantification and Design of Experiments
- History of interest, see e.g. McKay et al. (1979)
- The review:
- Random Sampling
- Stratified Sampling
- Latin Hypercube Sampling
- As approaches for Monte Carlo estimates
Random Sampling
Let the input values \(\mathbf{ x}_1, \dots, \mathbf{ x}_n\) be a random sample from \(f(\mathbf{ x})\). This method of sampling is perhaps the most obvious, and an entire body of statistical literature may be used in making inferences regarding the distribution of \(Y(t)\).
Stratified Sampling
Using stratified sampling, all areas of the sample space of \(\mathbf{ x}\) are represented by input values. Let the sample space \(S\) of \(\mathbf{ x}\) be partitioned into \(I\) disjoint strata \(S_t\). Let \(\pi = P(\mathbf{ x}\in S_i)\) represent the size of \(S_i\). Obtain a random sample \(\mathbf{ x}_{ij}\), \(j = 1, \dots, n\) from \(S_i\). Then of course the \(n_i\) sum to \(n\). If \(I = 1\), we have random sampling over the entire sample space.
Latin Hypercube Sampling
The same reasoning that led to stratified sampling, ensuring that all portions of \(S\) were sampled, could lead further. If we wish to ensure also that each of the input variables \(\mathbf{ x}_k\) has all portions of its distribution represented by input values, we can divide the range of each \(\mathbf{ x}_k\) into \(n\) strata of equal marginal probability \(1/n\), and sample once from each stratum. Let this sample be \(\mathbf{ x}_{kj}\), \(j = 1, \dots, n\). These form the \(\mathbf{ x}_k\) component, \(k = 1, \dots , K\), in \(\mathbf{ x}_i\), \(i = 1, \dots, n\). The components of the various \(\mathbf{ x}_k\)’s are matched at random. This method of selecting input values is an extension of quota sampling (Steinberg 1963), and can be viewed as a \(K\)-dimensional extension of Latin square sampling (Raj 1968).
Generative AI for Emulation
- Modern ML models can learn from simulation histories
- Example: GraphCast
for weather prediction
- Trained on archived weather forecasts
- Faster than traditional numerical simulation
- Maintains high accuracy
- Key requirement: Large archive of simulation runs
GraphCast Forecast
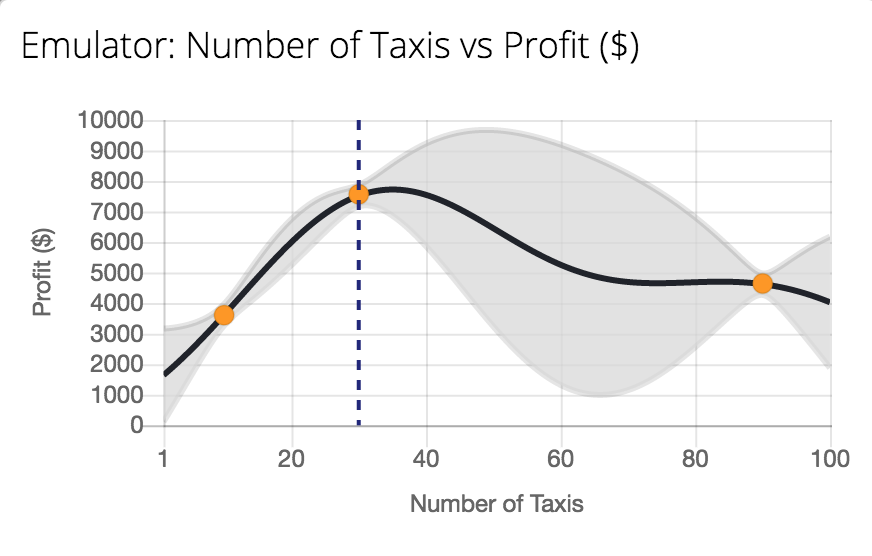
Emukit Playground
Work Leah Hirst, Software Engineering Intern and Cliff McCollum.
Tutorial on emulation.
Emukit Playground
Emukit Playground
Emukit
Emukit
Emukit
- Led by: Javier Gonzalez and Andrei Paleyes
- Team: Mark Pullin, Maren Mahsereci, Alex Gessner, Aaron Klein, Henry Moss and David-Elias Künstle.
- Management: Cliff McCollum & Neil
- Available on Github
- Example sensitivity notebook, documentation https://emukit.readthedocs.io/en/latest/
Modular Design
Introduce your own surrogate models.
To building your own model see this notebook.
Structure
- loop
- model
- candidate point calculator
- acquisition
- acquisition optimizer
- user function
- model updater
- stopping condition
Emukit Vision
Emukit and Emulation
Methods
- The different methods: Bayesian optimization, experimental design.
Models
- The probabilistic model that will be used to emulate. Emukit doesn’t define these, the user brings their own.
Tasks
- Still in development: High level goals that owners of the process/simulator might be actually interested in. Examples: measure quality of a simulator, explain complex system behavior.
Structure
while stopping condition is not met:
optimize acquisition function
evaluate user function
update model with new observationLoop
- An abstract class where the different components come together.
Model
- The surrogate model or emulator, often a Gaussian process.
Candidate Point Calculator
- The routine that combines acquisition with optimizer to compute the next candidate point (or points).
Acquisition
- Our acquisition function: in Bayesian Optimization, this might be Expected Improvement.
Acquisition Optimizer
- The optimization routine we use to optimize the acquisition function. (often this is a non-linear optimizer like L-BFGS (Byrd et al., 1995))
User Function
- The function we’re trying to reason about.
Model Updater
- How to update our surrogate model when we have new training data.
Stopping Condition
- How to decide when to stop our cycle of data acquisition from the target function.
Emukit Tutorial
Emukit Overview Summary
- Multi-fidelity emulation: build surrogate models for multiple sources of information;
- Bayesian optimisation: optimise physical experiments and tune parameters ML algorithms;
- Experimental design/Active learning: design experiments and perform active learning with ML models;
- Sensitivity analysis: analyse the influence of inputs on the outputs
- Bayesian quadrature: compute integrals of functions that are expensive to evaluate.
Model Free Experimental Design
Design of Experiments in Python
https://pythonhosted.org/pyDOE/
Experimental Design in Emukit
Forrester Function
Alex Forrester
The Forrester Function
\[ f(x) = (6x-2)^2\sin(12 x-4). \]
Initial Design
The Model
The Acquisition Function
Uncertainty Sampling
\[ a_{US}(\mathbf{ x}) = \sigma^2(\mathbf{ x}). \]
Integrated Variance Reduction
\[ \begin{align*} a_{\text{IVR}} & = \int_{\mathbb{X}}[\sigma^2(\mathbf{ x}') - \sigma^2(\mathbf{ x}'; \mathbf{ x})]\text{d}\mathbf{ x}' \\ & \approx \frac{1}{\# \text{samples}}\sum_i^{\# \text{samples}}[\sigma^2(\mathbf{ x}_i) - \sigma^2(\mathbf{ x}_i; \mathbf{ x})]. \end{align*} \]
\[ a_{LCB} \approx \frac{1}{\# \text{samples}}\sum_i^{\# \text{samples}}\frac{k^2(\mathbf{ x}_i, \mathbf{ x})}{\sigma^2(\mathbf{ x})}. \]
Evaluating the objective function
Add the New Point
The Model is Updated
Emukit’s Experimental Design Interface
Conclusions
- Emukit software.
- Example around experimental design.
- Sequential decision making with acquisiton functions.
- Generalizes from the BayesOpt process
(e.g.
GPyOpt)
Thanks!
book: The Atomic Human
twitter: @lawrennd
podcast: The Talking Machines
newspaper: Guardian Profile Page
blog posts: