Making sense of messy force spectroscopy data
Marie Synakewicz
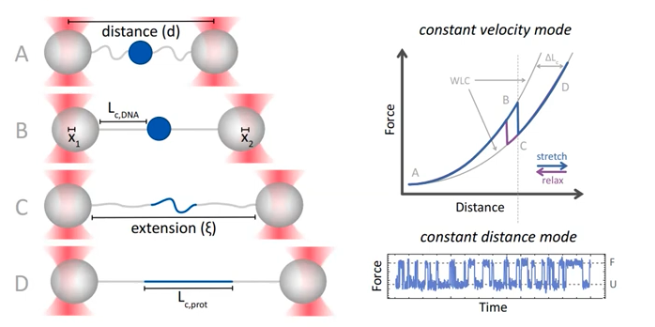
Abstract: All processes of life depend on the correct functioning of biomolecules. To investigate the biochemical and biophysical properties of biomolecules and how they are affected by changes in environment or by disease-causing mutations, scientists challenge them with chemicals, temperature, pressure or force. Here, we will focus on force spectroscopy, a type of experiment in which piconewton forces can be applied to single molecules. The first bottleneck that scientists encounter is that of setting up the experiment and acquiring the data. However, if they are really unlucky, there can be a second bottleneck: messy data that nobody knows what to do with! In the world of single-molecule force spectroscopy, such data usually signifies the end of a project. But what if we found new methods of analysing the data that could potentially be combined with biophysical models to help us understand the system under investigation? In this session, we will first cover a few basics on proteins, optical tweezers and the data that is produced, before discussing examples of ‘nice’ and ‘messy’ data, the inherent limitations and the necessary pre-processing.
links and publications:
- G. Žoldák and M. Rief. Force as a single molecule probe of multidimensional protein energy landscapes. Current Opinion in Structural Biology, 23(1):48–57, 2013.
- K. A. Dill and J. L. MacCallum. The protein-folding problem, 50 years on. Science, 338:1042–1046, 2012.

