Special Topics: Gaussian Processes
Review
- Last week: Logistic Regression and Generalised Linear Models
- Introduced link functions and different transformations.
- Showed examples in classification and mentioned possibilities for disease rate models.
- This week:
- Gaussian Processes: non parametric Bayesian modelling
Generalized Linear Models
- Logistic regression is a generalized linear model
- Prediction function that is linear in parameters \[ \log \frac{p(\mathbf{ x})}{1-p(\mathbf{ x})} = f(\mathbf{ x}; \mathbf{ w}) \]
- Where \(f(\cdot)\) is linear in parameters.
Basis Functions
Linear models have the form \[ f(\mathbf{ x}) = \mathbf{ w}^\top \boldsymbol{ \phi}(\mathbf{ x}). \]
Gaussian processes are related to generalized linear models.
Gaussian Processes
- Basis function models give non-linear predictions.
- Need to choose number and location of basis functions.
- Gaussian processes is a general framework (basis functions special case)
- Within the framework you can consider models with infinite basis functions.
\[ p(\mathbf{ y}|\mathbf{X}, \mathbf{ w}) = \prod_{i=1}^{n} p(y_i | \mathbf{ x}_i, \mathbf{ w}) \]
\[ \mathbf{ y}|\mathbf{X}\sim \mathcal{N}\left(\mathbf{m}(\mathbf{X}),\mathbf{K}(\mathbf{X})\right), \]
Sampling a Function
Multi-variate Gaussians
- We will consider a Gaussian with a particular structure of covariance matrix.
- Generate a single sample from this 25 dimensional Gaussian density, \[ \mathbf{ f}=\left[f_{1},f_{2}\dots f_{25}\right]. \]
- We will plot these points against their index.
Gaussian Distribution Sample
Sampling a Function from a Gaussian
Joint Density of \(f_1\) and \(f_2\)
Prediction of \(f_{2}\) from \(f_{1}\)
Uluru

Prediction with Correlated Gaussians
- Prediction of \(f_2\) from \(f_1\) requires conditional density.
- Conditional density is also Gaussian. \[ p(f_2|f_1) = \mathcal{N}\left(f_2|\frac{k_{1, 2}}{k_{1, 1}}f_1, k_{2, 2} - \frac{k_{1,2}^2}{k_{1,1}}\right) \] where covariance of joint density is given by \[ \mathbf{K}= \begin{bmatrix} k_{1, 1} & k_{1, 2}\\ k_{2, 1} & k_{2, 2}.\end{bmatrix} \]
Joint Density of \(f_1\) and \(f_8\)
Prediction of \(f_{8}\) from \(f_{1}\)
Details
- The single contour of the Gaussian density represents the joint distribution, \(p(f_1, f_8)\)
- We observe a value for \(f_1=-?\)
- Conditional density: \(p(f_8|f_1=?)\).
Prediction with Correlated Gaussians
Prediction of \(\mathbf{ f}_*\) from \(\mathbf{ f}\) requires multivariate conditional density.
Multivariate conditional density is also Gaussian.
\[ p(\mathbf{ f}_*|\mathbf{ f}) = {\mathcal{N}\left(\mathbf{ f}_*|\mathbf{K}_{*,\mathbf{ f}}\mathbf{K}_{\mathbf{ f},\mathbf{ f}}^{-1}\mathbf{ f},\mathbf{K}_{*,*}-\mathbf{K}_{*,\mathbf{ f}} \mathbf{K}_{\mathbf{ f},\mathbf{ f}}^{-1}\mathbf{K}_{\mathbf{ f},*}\right)} \] Here covariance of joint density is given by \[ \mathbf{K}= \begin{bmatrix} \mathbf{K}_{\mathbf{ f}, \mathbf{ f}} & \mathbf{K}_{*, \mathbf{ f}}\\ \mathbf{K}_{\mathbf{ f}, *} & \mathbf{K}_{*, *}\end{bmatrix} \]
Prediction with Correlated Gaussians
Prediction of \(\mathbf{ f}_*\) from \(\mathbf{ f}\) requires multivariate conditional density.
Multivariate conditional density is also Gaussian.
\[ p(\mathbf{ f}_*|\mathbf{ f}) = {\mathcal{N}\left(\mathbf{ f}_*|\boldsymbol{ \mu},\boldsymbol{ \Sigma}\right)} \] \[ \boldsymbol{ \mu}= \mathbf{K}_{*,\mathbf{ f}}\mathbf{K}_{\mathbf{ f},\mathbf{ f}}^{-1}\mathbf{ f} \] \[ \boldsymbol{ \Sigma}= \mathbf{K}_{*,*}-\mathbf{K}_{*,\mathbf{ f}} \mathbf{K}_{\mathbf{ f},\mathbf{ f}}^{-1}\mathbf{K}_{\mathbf{ f},*} \] Here covariance of joint density is given by \[ \mathbf{K}= \begin{bmatrix} \mathbf{K}_{\mathbf{ f}, \mathbf{ f}} & \mathbf{K}_{*, \mathbf{ f}}\\ \mathbf{K}_{\mathbf{ f}, *} & \mathbf{K}_{*, *}\end{bmatrix} \]
Marginal Likelihood
Sampling from the Prior
Weight Space View
\[ \mathbf{ w}\sim \mathcal{N}\left(\mathbf{0},\alpha\mathbf{I}\right), \]
\[ f(\mathbf{ x}) = \mathbf{ w}^\top \boldsymbol{ \phi}(\mathbf{ x}). \]
Function Space View
\[ \mathbf{ w}\sim \mathcal{N}\left(\mathbf{0},\alpha \mathbf{I}\right) \]
\[ \boldsymbol{ \Phi}= \begin{bmatrix}\boldsymbol{ \phi}(\mathbf{ x}_1) \\ \vdots \\ \boldsymbol{ \phi}(\mathbf{ x}_n)\end{bmatrix} \]
\[ \mathbf{ f}= \begin{bmatrix} f_1 \\ \vdots f_n\end{bmatrix} \]
\[ \mathbf{ f}= \boldsymbol{ \Phi}\mathbf{ w}. \]
\[ \mathbf{ f}\sim \mathcal{N}\left(\mathbf{0},\alpha \boldsymbol{ \Phi}\boldsymbol{ \Phi}^\top\right). \]
\[ \mathbf{K}= \alpha \boldsymbol{ \Phi}\boldsymbol{ \Phi}^\top. \]
K = alpha*Phi_pred@Phi_pred.T
f_sample = np.random.multivariate_normal(mean=np.zeros(x_pred.size), cov=K)\[ \mathbf{ y}= \mathbf{ f}+ \boldsymbol{\epsilon} \]
\[ \epsilon \sim \mathcal{N}\left(\mathbf{0},\sigma^2\mathbf{I}\right). \]
\[ \mathbf{ y}\sim \mathcal{N}\left(\mathbf{0},\boldsymbol{ \Phi}\boldsymbol{ \Phi}^\top +\sigma^2\mathbf{I}\right). \]
Non-degenerate Gaussian Processes
- This process is degenerate.
- Covariance function is of rank at most \(h\).
- As \(n\rightarrow \infty\), covariance matrix is not full rank.
- Leading to \(\det{\mathbf{K}} = 0\)
Infinite Networks
- In ML Radford Neal (Neal, 1994) asked “what would happen if you took \(h\rightarrow \infty\)?”
Roughly Speaking
- Instead of \[ \begin{align*} k_f\left(\mathbf{ x}_i, \mathbf{ x}_j\right) & = \alpha \boldsymbol{ \phi}\left(\mathbf{W}_1, \mathbf{ x}_i\right)^\top \boldsymbol{ \phi}\left(\mathbf{W}_1, \mathbf{ x}_j\right)\\ & = \alpha \sum_k \phi\left(\mathbf{ w}^{(1)}_k, \mathbf{ x}_i\right) \phi\left(\mathbf{ w}^{(1)}_k, \mathbf{ x}_j\right) \end{align*} \]
- Sample infinitely many from a prior density, \(p(\mathbf{ w}^{(1)})\), \[ k_f\left(\mathbf{ x}_i, \mathbf{ x}_j\right) = \alpha \int \phi\left(\mathbf{ w}^{(1)}, \mathbf{ x}_i\right) \phi\left(\mathbf{ w}^{(1)}, \mathbf{ x}_j\right) p(\mathbf{ w}^{(1)}) \text{d}\mathbf{ w}^{(1)} \]
- Also applies for non-Gaussian \(p(\mathbf{ w}^{(1)})\) because of the central limit theorem.
Simple Probabilistic Program
If \[ \begin{align*} \mathbf{ w}^{(1)} & \sim p(\cdot)\\ \phi_i & = \phi\left(\mathbf{ w}^{(1)}, \mathbf{ x}_i\right), \end{align*} \] has finite variance.
Then taking number of hidden units to infinity, is also a Gaussian process.
Further Reading
Chapter 2 of Neal’s thesis (Neal, 1994)
Rest of Neal’s thesis. (Neal, 1994)
David MacKay’s PhD thesis (MacKay, 1992)
Gaussian Process
\[ k(\mathbf{ x}, \mathbf{ x}^\prime) = \alpha \exp\left( -\frac{\left\Vert \mathbf{ x}-\mathbf{ x}^\prime\right\Vert^2}{2\ell^2}\right), \]
\[ \left\Vert\mathbf{ x}- \mathbf{ x}^\prime\right\Vert^2 = (\mathbf{ x}- \mathbf{ x}^\prime)^\top (\mathbf{ x}- \mathbf{ x}^\prime) \]

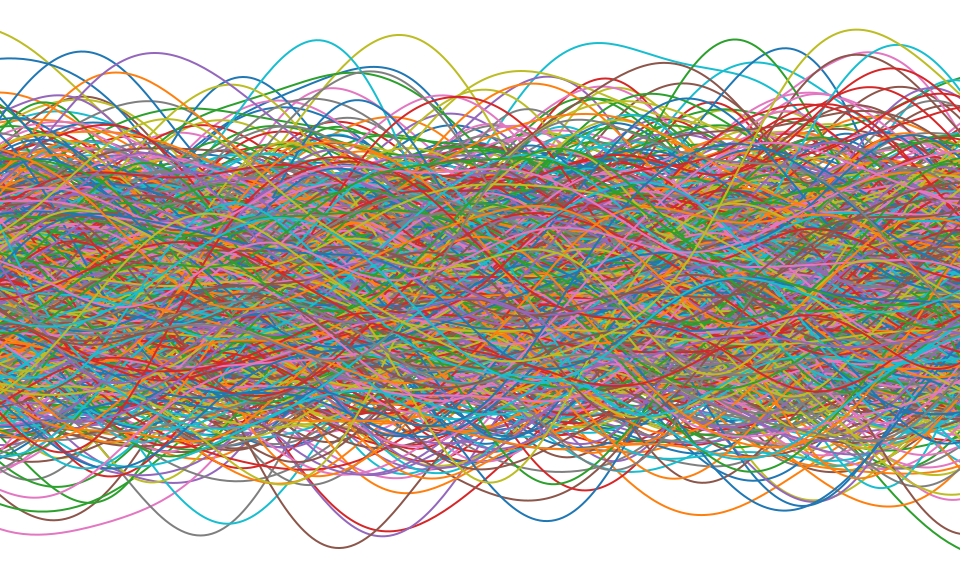
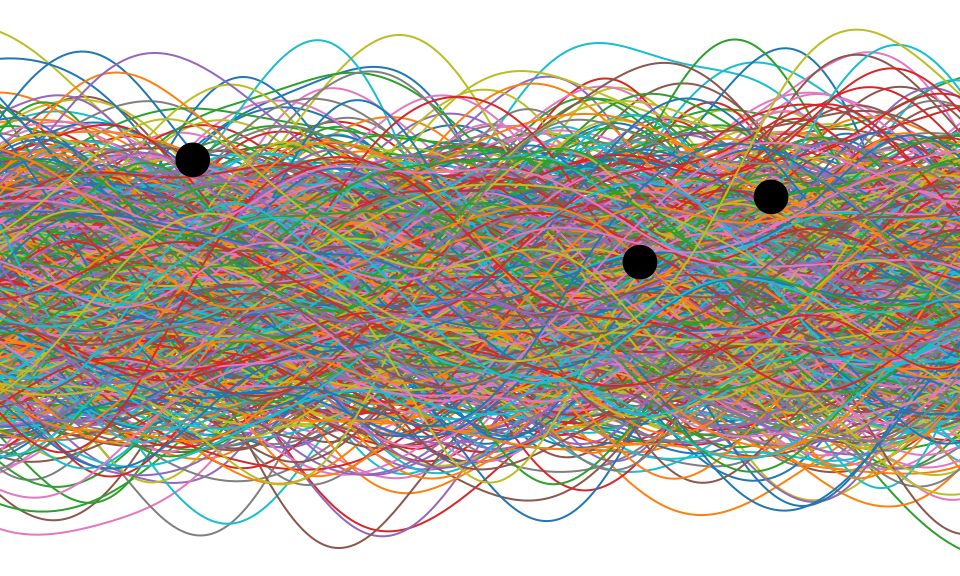


Gaussian Process
\[ p(\mathbf{ y}|\mathbf{X}) = \frac{1}{(2\pi)^{\frac{n}{2}}|\mathbf{K}|^{\frac{1}{2}}} \exp\left(-\frac{1}{2}\mathbf{ y}^\top \left(\mathbf{K}+\sigma^2 \mathbf{I}\right)^{-1}\mathbf{ y}\right) \]
\[ E(\boldsymbol{\theta}) = \frac{1}{2} \log |\mathbf{K}| + \frac{1}{2} \mathbf{ y}^\top \left(\mathbf{K}+ \sigma^2\mathbf{I}\right)^{-1}\mathbf{ y} \]
Making Predictions
\[ \begin{bmatrix}\mathbf{ f}\\ \mathbf{ f}^*\end{bmatrix} \sim \mathcal{N}\left(\mathbf{0},\begin{bmatrix} \mathbf{K}& \mathbf{K}_\ast \\ \mathbf{K}_\ast^\top & \mathbf{K}_{\ast,\ast}\end{bmatrix}\right) \]
\[ \begin{bmatrix} \mathbf{K}& \mathbf{K}_\ast \\ \mathbf{K}_\ast^\top & \mathbf{K}_{\ast,\ast}\end{bmatrix} \]
The Importance of the Covariance Function
\[ \boldsymbol{ \mu}_f= \mathbf{A}^\top \mathbf{ y}, \]
Improving the Numerics
In practice we shouldn’t be using matrix inverse directly to solve the GP system. One more stable way is to compute the Cholesky decomposition of the kernel matrix. The log determinant of the covariance can also be derived from the Cholesky decomposition.
Capacity Control
Gradients of the Likelihood
Overall Process Scale
Capacity Control and Data Fit
Learning Covariance Parameters
Can we determine covariance parameters from the data?
\[ \mathcal{N}\left(\mathbf{ y}|\mathbf{0},\mathbf{K}\right)=\frac{1}{(2\pi)^\frac{n}{2}{\det{\mathbf{K}}^{\frac{1}{2}}}}{\exp\left(-\frac{\mathbf{ y}^{\top}\mathbf{K}^{-1}\mathbf{ y}}{2}\right)} \]
\[ \begin{aligned} \mathcal{N}\left(\mathbf{ y}|\mathbf{0},\mathbf{K}\right)=\frac{1}{(2\pi)^\frac{n}{2}\color{yellow}{\det{\mathbf{K}}^{\frac{1}{2}}}}\color{cyan}{\exp\left(-\frac{\mathbf{ y}^{\top}\mathbf{K}^{-1}\mathbf{ y}}{2}\right)} \end{aligned} \]
\[ \begin{aligned} \log \mathcal{N}\left(\mathbf{ y}|\mathbf{0},\mathbf{K}\right)=&\color{yellow}{-\frac{1}{2}\log\det{\mathbf{K}}}\color{cyan}{-\frac{\mathbf{ y}^{\top}\mathbf{K}^{-1}\mathbf{ y}}{2}} \\ &-\frac{n}{2}\log2\pi \end{aligned} \]
\[ E(\boldsymbol{ \theta}) = \color{yellow}{\frac{1}{2}\log\det{\mathbf{K}}} + \color{cyan}{\frac{\mathbf{ y}^{\top}\mathbf{K}^{-1}\mathbf{ y}}{2}} \]
Capacity Control through the Determinant
The parameters are inside the covariance function (matrix). \[k_{i, j} = k(\mathbf{ x}_i, \mathbf{ x}_j; \boldsymbol{ \theta})\]
Eigendecomposition of Covariance
\[\mathbf{K}= \mathbf{R}\boldsymbol{ \Lambda}^2 \mathbf{R}^\top\]

|
\(\boldsymbol{ \Lambda}\) represents distance on axes. \(\mathbf{R}\) gives rotation. |
Eigendecomposition of Covariance
- \(\boldsymbol{ \Lambda}\) is diagonal, \(\mathbf{R}^\top\mathbf{R}= \mathbf{I}\).
- Useful representation since \(\det{\mathbf{K}} = \det{\boldsymbol{ \Lambda}^2} = \det{\boldsymbol{ \Lambda}}^2\).
Capacity control: \(\color{yellow}{\log \det{\mathbf{K}}}\)
Quadratic Data Fit
Data Fit: \(\color{cyan}{\frac{\mathbf{ y}^\top\mathbf{K}^{-1}\mathbf{ y}}{2}}\)
\[E(\boldsymbol{ \theta}) = \color{yellow}{\frac{1}{2}\log\det{\mathbf{K}}}+\color{cyan}{\frac{\mathbf{ y}^{\top}\mathbf{K}^{-1}\mathbf{ y}}{2}}\]
Data Fit Term
Exponentiated Quadratic Covariance

|
Olympic Marathon Data
|

|
Olympic Marathon Data
Alan Turing

|

|
Probability Winning Olympics?
- He was a formidable Marathon runner.
- In 1946 he ran a time 2 hours 46 minutes.
- That’s a pace of 3.95 min/km.
- What is the probability he would have won an Olympics if one had been held in 1946?
Gaussian Process Fit
Olympic Marathon Data GP
Della Gatta Gene Data
- Given given expression levels in the form of a time series from Della Gatta et al. (2008).
Della Gatta Gene Data
Gene Expression Example
- Want to detect if a gene is expressed or not, fit a GP to each gene Kalaitzis and Lawrence (2011).

TP53 Gene Data GP
TP53 Gene Data GP
TP53 Gene Data GP
Multiple Optima
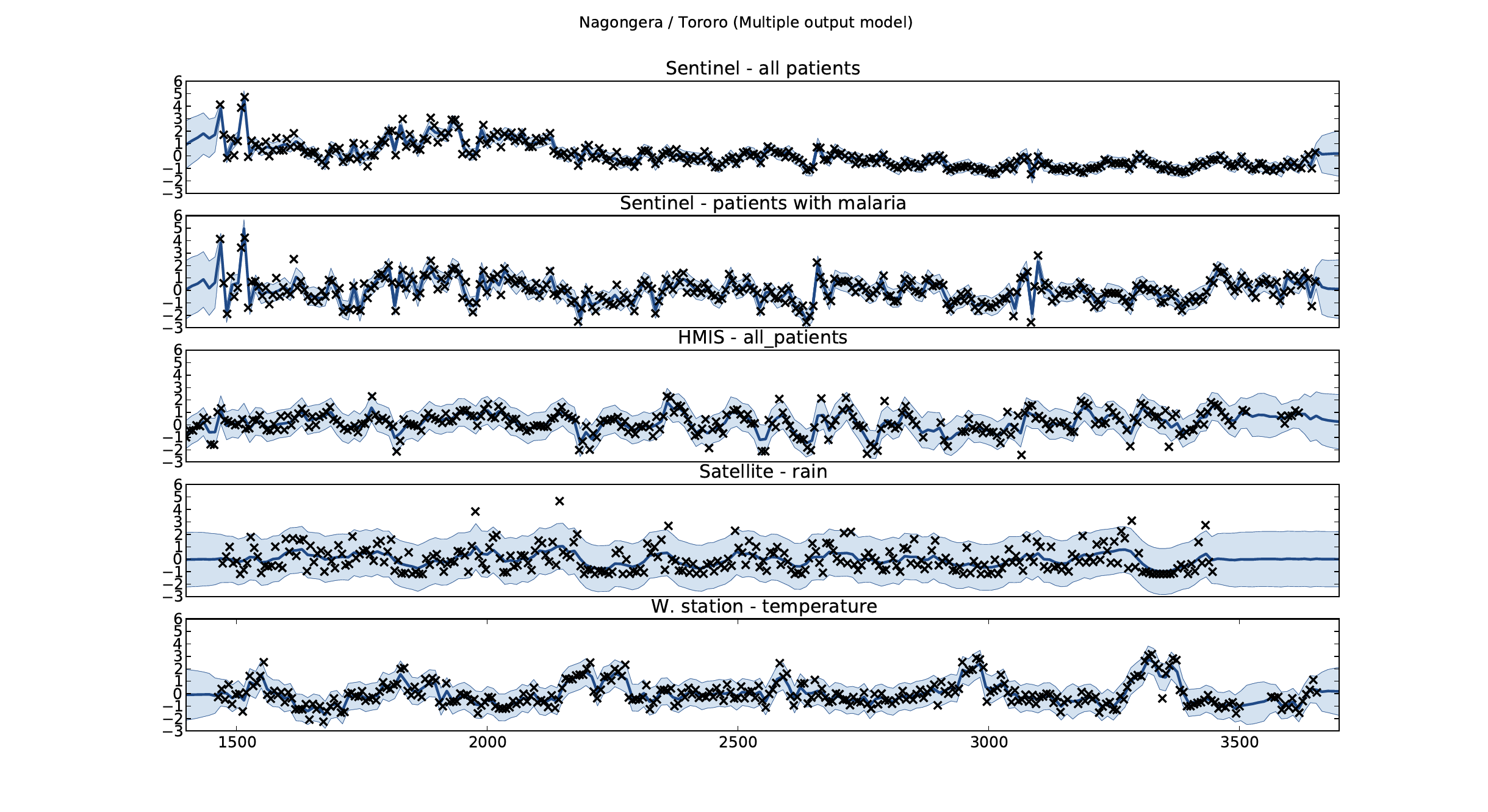
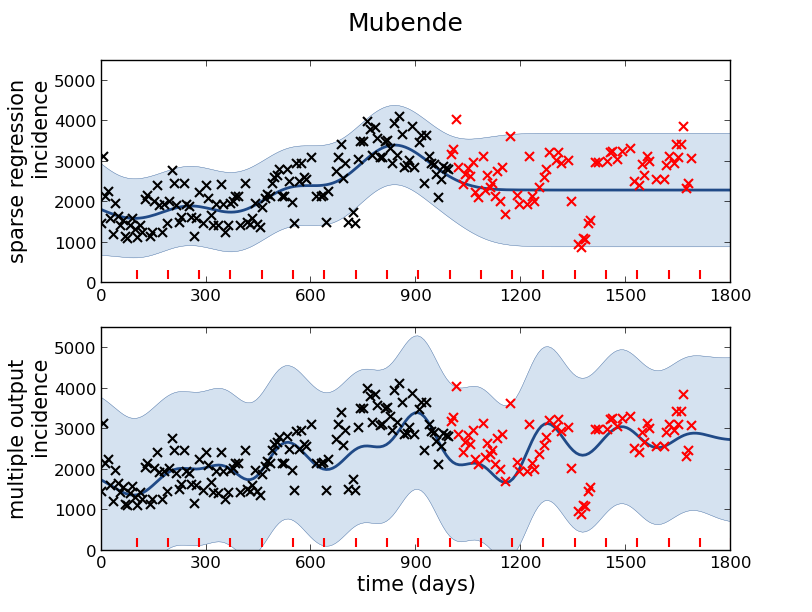
Example: Prediction of Malaria Incidence in Uganda
- Work with Ricardo Andrade Pacheco, John Quinn and Martin Mubangizi (Makerere University, Uganda)
- See AI-DEV Group.
- See UN Global Pulse Disease Outbreaks Site
Malaria Prediction in Uganda
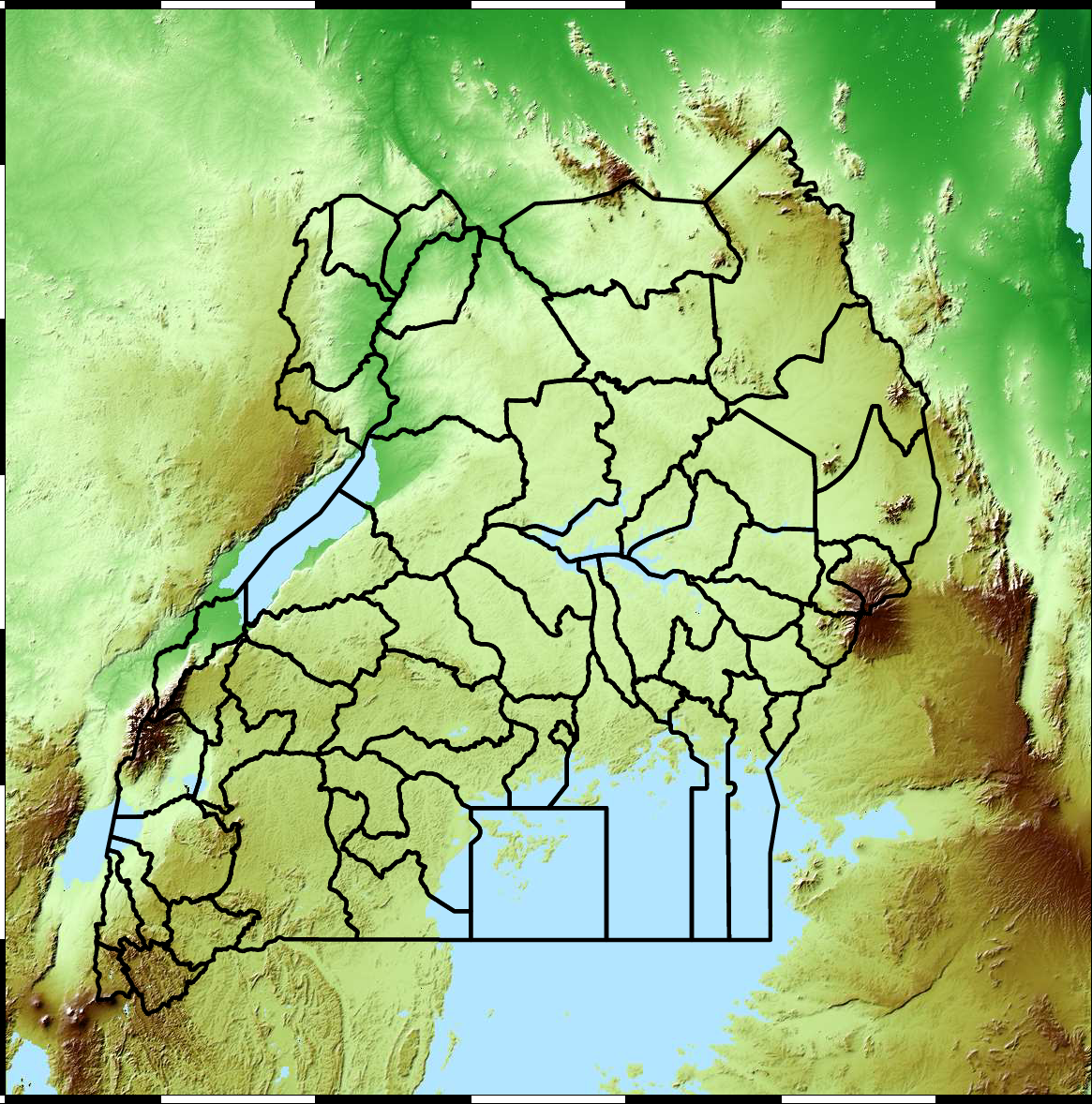
Kapchorwa District
Tororo District
Malaria Prediction in Nagongera (Sentinel Site)
Mubende District
Malaria Prediction in Uganda
GP School at Makerere

Kabarole District
Early Warning System

Early Warning Systems

Additive Covariance

|
Gelman Book

|
|
Basis Function Covariance

|
Brownian Covariance

|
MLP Covariance

|
GPSS: Gaussian Process Summer School
|
|
GPy: A Gaussian Process Framework in Python

GPy: A Gaussian Process Framework in Python
- BSD Licensed software base.
- Wide availability of libraries, ‘modern’ scripting language.
- Allows us to set projects to undergraduates in Comp Sci that use GPs.
- Available through GitHub https://github.com/SheffieldML/GPy
- Reproducible Research with Jupyter Notebook.
Features
- Probabilistic-style programming (specify the model, not the algorithm).
- Non-Gaussian likelihoods.
- Multivariate outputs.
- Dimensionality reduction.
- Approximations for large data sets.
Thanks!
twitter: @lawrennd
podcast: The Talking Machines
newspaper: Guardian Profile Page
blog posts: