Visualisation I: Discrete and Continuous Latent Variables
LT2, William Gates Building
Visualization and Human Perception
- Human visual system is our highest bandwidth connection to the world
- Optic tract: ~8.75 million bits/second
- Verbal communication: only ~2,000 bits/minute
- Active sensing through rapid eye movements (saccades)
- Not passive reception
- Actively construct understanding
- Hundreds of samples per second
Behind the Eye

Visualization and Human Perception
- Visualization is powerful for communication
- But can be vulnerable to manipulation
- Similar to social media algorithms
- Misleading visualizations can deceive
- Can hijack natural visual processing

Part 1: Discrete Latent Variables
Clustering
Clustering
- Common approach for grouping data points
- Assigns data points to discrete groups
- Examples include:
- Animal classification
- Political affiliation grouping
Clustering vs Vector Quantisation
- Clustering expects gaps between groups in data density
- Vector quantization may not require density gaps
- For practical purposes, both involve:
- Allocating points to groups
- Determining optimal number of groups
\(k\)-means Clustering
- Simple iterative clustering algorithm
- Key steps:
- Initialize with random centers
- Assign points to nearest center
- Update centers as cluster means
- Repeat until stable
Objective Function
Minimizes sum of squared distances: \[ E=\sum_{j=1}^K \sum_{i\ \text{allocated to}\ j} \left(\mathbf{ y}_{i, :} - \boldsymbol{ \mu}_{j, :}\right)^\top\left(\mathbf{ y}_{i, :} - \boldsymbol{ \mu}_{j, :}\right) \]
Solution not guaranteed to be global or unique
Represents a non-convex optimization problem
Task: associate data points with different labels.
Labels are not provided by humans.
Process is intuitive for humans - we do it naturally.
Platonic Ideals
- Greek philosopher Plato considered the concept of ideals
- The Platonic ideal bird is the most bird-like bird
- In clustering, we find these ideals as cluster centers
- Data points are allocated to their nearest center
Mathematical Formulation
- Represent objects as data vectors \(\mathbf{ x}_i\)
- Represent cluster centers as vectors \(\boldsymbol{ \mu}_j\)
- Define similarity/distance between objects and centers
- Distance function: \(d_{ij} = f(\mathbf{ x}_i, \boldsymbol{ \mu}_j)\)
Squared Distance
- Common choice: squared distance \[ d_{ij} = (\mathbf{ x}_i - \boldsymbol{ \mu}_j)^2 \]
- Goal: find centers close to many data points
Objective Function
- Given similarity measure, need number of cluster centers, \(K\).
- Find their location by allocating each center to a sub-set of the points and minimizing the sum of the squared errors, \[ E(\mathbf{M}) = \sum_{i \in \mathbf{i}_j} (\mathbf{ x}_i - \boldsymbol{ \mu}_j)^2 \] here \(\mathbf{i}_j\) is all indices of data points allocated to the \(j\)th center.
\(k\)-Means Clustering
- \(k\)-means clustering is simple and quick to implement.
- Very initialisation sensitive.
Initialisation
- Initialisation is the process of selecting a starting set of parameters.
- Optimisation result can depend on the starting point.
- For \(k\)-means clustering you need to choose an initial set of centers.
- Optimisation surface has many local optima, algorithm gets stuck in ones near initialisation.
\(k\)-Means Clustering
Clustering with the \(k\)-means clustering algorithm.
\(k\)-Means Clustering
\(k\)-means clustering by Alex Ihler
Hierarchical Clustering
- Form taxonomies of the cluster centers
- Like humans apply to animals, to form phylogenies
- Builds a tree structure showing relationships between data points
- Two main approaches:
- Agglomerative (bottom-up): Start with individual points and merge
- Divisive (top-down): Start with one cluster and split
Oil Flow Data
}
Phylogenetic Trees
- Hierarchical clustering of genetic sequence data
- Creates evolutionary trees showing species relationships
- Estimates common ancestors and mutation timelines
- Critical for tracking viral evolution and outbreaks
Product Clustering
- Hierarchical clustering for e-commerce products
- Creates product taxonomy trees
- Splits into nested categories (e.g. Electronics → Phones → Smartphones)
Hierarchical Clustering Challenge
- Many products belong in multiple clusters (e.g. running shoes are both ‘sporting goods’ and ‘clothing’)
- Tree structures are too rigid for natural categorization
- Human concept learning is more flexible:
- Forms overlapping categories
- Learns abstract rules
- Builds causal theories
Other Clustering Approaches
- Spectral clustering: Graph-based non-convex clustering
- Dirichlet process: Infinite, non-parametric clustering
Part 2: Continuous Latent Variables
High Dimensional Data
- USPS Data Set Handwritten Digit
- 3648 dimensions (64 rows, 57 columns)
- Space contains much more than just this digit.
USPS Samples




- Even if we sample every nanonsecond from now until end of universe you won’t see original six!
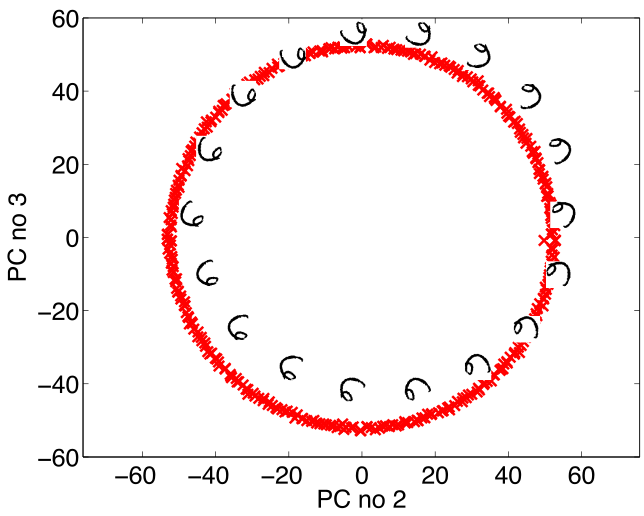
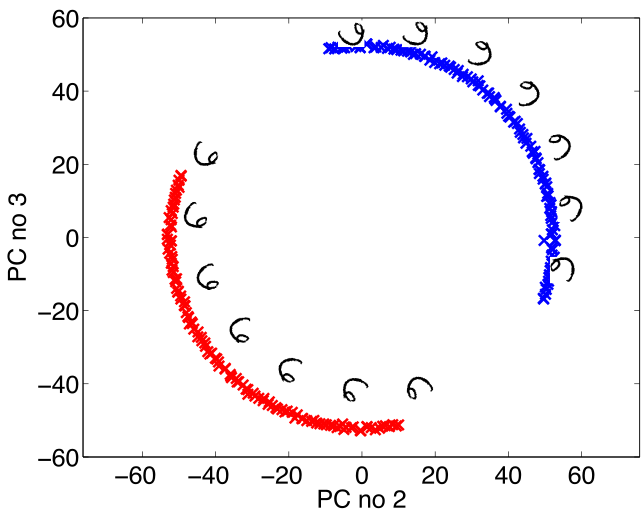
Simple Model of Digit
- Rotate a prototype





|
|
Low Dimensional Manifolds
- Pure rotation is too simple
- In practice data may undergo several distortions.
- For high dimensional data with structure:
- We expect fewer distortions than dimensions;
- Therefore we expect the data to live on a lower dimensional manifold.
- Conclusion: Deal with high dimensional data by looking for a lower dimensional non-linear embedding.
High Dimensional Data Effects
- High dimensional spaces behave very differently from our 3D intuitions
- Two key effects:
- Data moves to a “shell” at one standard deviation from mean
- Distances between points become constant
- Let’s see this experimentally
Pairwise Distances in High-D Gaussian Data
Pairwise Distances in High-D Gaussian Data
- Plot shows pairwise distances in high-D Gaussian data
- Red line: theoretical gamma distribution
- Notice tight concentration of distances
Structured High Dimensional Data
- What about data with underlying structure?
- Let’s create data that lies on a 2D manifold
- Embed it in 1000D space
Pairwise Distances in Structured High-D Data
Pairwise Distances in Structured High-D Data
- Distance distribution differs from pure high-D case
- Matches 2D theoretical curve better than 1000D
- Real data often has low intrinsic dimensionality
- This is why PCA and other dimension reduction works!
High Dimensional Effects in Real Data
Oil Flow Data
Implications for Dimensionality Reduction
Latent Variables and Dimensionality Reduction
- Real data often has lower intrinsic dimensionality than measurements
- Examples:
- Motion capture: Many coordinates but few degrees of freedom
- Genetic data: Thousands of genes controlled by few regulators
- Images: Millions of pixels but simpler underlying structure
Latent Variable Example
Latent Variable Example
- Example shows 2D data described by 1D latent variable
- Left: Data in original 2D space
- Right: Same data represented by single latent variable \(z\)
- Goal: Find these simpler underlying representations
Latent Variables
Your Personality
Factor Analysis Model
\[ \mathbf{ y}= \mathbf{f}(\mathbf{ z}) + \boldsymbol{ \epsilon}, \]
\[ \mathbf{f}(\mathbf{ z}) = \mathbf{W}\mathbf{ z} \]
Closely Related to Linear Regression
\[ \mathbf{f}(\mathbf{ z}) = \begin{bmatrix} f_1(\mathbf{ z}) \\ f_2(\mathbf{ z}) \\ \vdots \\ f_p(\mathbf{ z})\end{bmatrix} \]
\[ f_j(\mathbf{ z}) = \mathbf{ w}_{j, :}^\top \mathbf{ z}, \]
\[ \epsilon_j \sim \mathcal{N}\left(0,\sigma^2_j\right). \]
Data Representation
\[ \mathbf{Y} = \begin{bmatrix} \mathbf{ y}_{1, :}^\top \\ \mathbf{ y}_{2, :}^\top \\ \vdots \\ \mathbf{ y}_{n, :}^\top\end{bmatrix}, \]
\[ \mathbf{F} = \mathbf{Z}\mathbf{W}^\top, \]
Latent Variables vs Linear Regression
\[ x_{i,j} \sim \mathcal{N}\left(0,1\right), \] and we can write the density governing the latent variable associated with a single point as, \[ \mathbf{ z}_{i, :} \sim \mathcal{N}\left(\mathbf{0},\mathbf{I}\right). \]
\[ \mathbf{f}_{i, :} = \mathbf{f}(\mathbf{ z}_{i, :}) = \mathbf{W}\mathbf{ z}_{i, :} \]
\[ \mathbf{f}_{i, :} \sim \mathcal{N}\left(\mathbf{0},\mathbf{W}\mathbf{W}^\top\right) \]
Data Distribution
\[ \mathbf{ y}_{i, :} = \sim \mathcal{N}\left(\mathbf{0},\mathbf{W}\mathbf{W}^\top + \boldsymbol{\Sigma}\right) \]
\[ \boldsymbol{\Sigma} = \begin{bmatrix}\sigma^2_{1} & 0 & 0 & 0\\ 0 & \sigma^2_{2} & 0 & 0\\ 0 & 0 & \ddots & 0\\ 0 & 0 & 0 & \sigma^2_p\end{bmatrix}. \]
Mean Vector
\[ \mathbf{ y}_{i, :} = \mathbf{W}\mathbf{ z}_{i, :} + \boldsymbol{ \mu}+ \boldsymbol{ \epsilon}_{i, :} \]
\[ \boldsymbol{ \mu}= \frac{1}{n} \sum_{i=1}^n \mathbf{ y}_{i, :}, \] \(\mathbf{C}= \mathbf{W}\mathbf{W}^\top + \boldsymbol{\Sigma}\)
Principal Component Analysis
Hotelling (1933) took \(\sigma^2_i \rightarrow 0\) so \[ \mathbf{ y}_{i, :} \sim \lim_{\sigma^2 \rightarrow 0} \mathcal{N}\left(\mathbf{0},\mathbf{W}\mathbf{W}^\top + \sigma^2 \mathbf{I}\right). \]
Degenerate Covariance
\[ p(\mathbf{ y}_{i, :}|\mathbf{W}) = \lim_{\sigma^2 \rightarrow 0} \frac{1}{(2\pi)^\frac{p}{2} |\mathbf{W}\mathbf{W}^\top + \sigma^2 \mathbf{I}|^{\frac{1}{2}}} \exp\left(-\frac{1}{2}\mathbf{ y}_{i, :}\left[\mathbf{W}\mathbf{W}^\top+ \sigma^2 \mathbf{I}\right]^{-1}\mathbf{ y}_{i, :}\right), \]
Computation of the Marginal Likelihood
\[ \mathbf{ y}_{i,:}=\mathbf{W}\mathbf{ z}_{i,:}+\boldsymbol{ \epsilon}_{i,:},\quad \mathbf{ z}_{i,:} \sim \mathcal{N}\left(\mathbf{0},\mathbf{I}\right), \quad \boldsymbol{ \epsilon}_{i,:} \sim \mathcal{N}\left(\mathbf{0},\sigma^{2}\mathbf{I}\right) \]
\[ \mathbf{W}\mathbf{ z}_{i,:} \sim \mathcal{N}\left(\mathbf{0},\mathbf{W}\mathbf{W}^\top\right) \]
\[ \mathbf{W}\mathbf{ z}_{i, :} + \boldsymbol{ \epsilon}_{i, :} \sim \mathcal{N}\left(\mathbf{0},\mathbf{W}\mathbf{W}^\top + \sigma^2 \mathbf{I}\right) \]
Linear Latent Variable Model II
Probabilistic PCA Max. Likelihood Soln (Tipping and Bishop (1999))

\[p\left(\mathbf{Y}|\mathbf{W}\right)=\prod_{i=1}^{n}\mathcal{N}\left(\mathbf{ y}_{i, :}|\mathbf{0},\mathbf{W}\mathbf{W}^{\top}+\sigma^{2}\mathbf{I}\right)\]
Linear Latent Variable Model II
Probabilistic PCA Max. Likelihood Soln (Tipping and Bishop (1999)) \[ p\left(\mathbf{Y}|\mathbf{W}\right)=\prod_{i=1}^{n}\mathcal{N}\left(\mathbf{ y}_{i,:}|\mathbf{0},\mathbf{C}\right),\quad \mathbf{C}=\mathbf{W}\mathbf{W}^{\top}+\sigma^{2}\mathbf{I} \] \[ \log p\left(\mathbf{Y}|\mathbf{W}\right)=-\frac{n}{2}\log\left|\mathbf{C}\right|-\frac{1}{2}\text{tr}\left(\mathbf{C}^{-1}\mathbf{Y}^{\top}\mathbf{Y}\right)+\text{const.} \] If \(\mathbf{U}_{q}\) are first \(q\) principal eigenvectors of \(n^{-1}\mathbf{Y}^{\top}\mathbf{Y}\) and the corresponding eigenvalues are \(\boldsymbol{\Lambda}_{q}\), \[ \mathbf{W}=\mathbf{U}_{q}\mathbf{L}\mathbf{R}^{\top},\quad\mathbf{L}=\left(\boldsymbol{\Lambda}_{q}-\sigma^{2}\mathbf{I}\right)^{\frac{1}{2}} \] where \(\mathbf{R}\) is an arbitrary rotation matrix.
Thanks!
book: The Atomic Human
twitter: @lawrennd
The Atomic Human pages bandwidth, communication 10-12,16,21,29,31,34,38,41,44,65-67,76,81,90-91,104,115,149,196,214,216,235,237-238,302,334 , MacKay, Donald 227-228,230-237,267-270, optic nerve/tract 205,235, O’Regan, Kevin 236-240,250,259,262-263,297,299, saccade 236,238,259-260,297,301, visual system/visual cortex 204-206,209,235-239,249-250,255,259,260,268-270,281,294,297,301,324,330.
podcast: The Talking Machines
newspaper: Guardian Profile Page
blog posts: